Mitochondria-rough-ER contacts in the liver regulate systemic lipid homeostasis
In this work, we studied mitochondria-rER contacts in vivo by serial section electron tomography (SSET) and 3D reconstruction analysis of cryo-fixed mouse tissue samples. We characterized this inter-organelle association as mitochondria tightly wrapped by sheets of curved rER (wrappER). Further, we used multi-omics and genetic approaches to obtain evidence that the wrappER is a distinct intracellular compartment and demonstrate the importance of wrappER-mitochondria contacts for very-low-density lipoprotein (VLDL) secretion from the liver, thereby connecting intracellular and systemic control mechanisms for lipid metabolism and homeostasis.
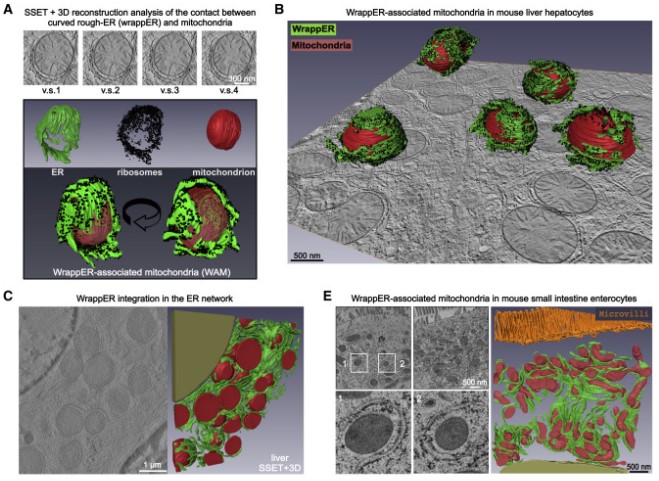
Contacts between organelles create microdomains that play major roles in regulating key intracellular activities and signaling pathways, but whether they also regulate systemic functions remains unknown. Here, we report the ultrastructural organization and dynamics of the inter-organellar contact established by sheets of curved rough endoplasmic reticulum closely wrapped around the mitochondria (wrappER). To elucidate the in vivo function of this contact, mouse liver fractions enriched in wrappER-associated mitochondria are analyzed by transcriptomics, proteomics, and lipidomics. The biochemical signature of the wrappER points to a role in the biogenesis of very-low-density lipoproteins (VLDL). Altering wrappER-mitochondria contacts curtails VLDL secretion and increases hepatic fatty acids, lipid droplets, and neutral lipid content. Conversely, acute liver-specific ablation of Mttp, the most upstream regulator of VLDL biogenesis, recapitulates this hepatic dyslipidemia phenotype and promotes remodeling of the wrappER-mitochondria contact. The discovery that liver wrappER-mitochondria contacts participate in VLDL biology suggests an involvement of inter-organelle contacts in systemic lipid homeostasis.