Patterns of organelle ontogeny through a cell cycle revealed by whole-cell reconstructions using 3D electron microscopy
Louise Hughes, Samantha Borrett, Katie Towers, Tobias Starborg, Sue Vaughan
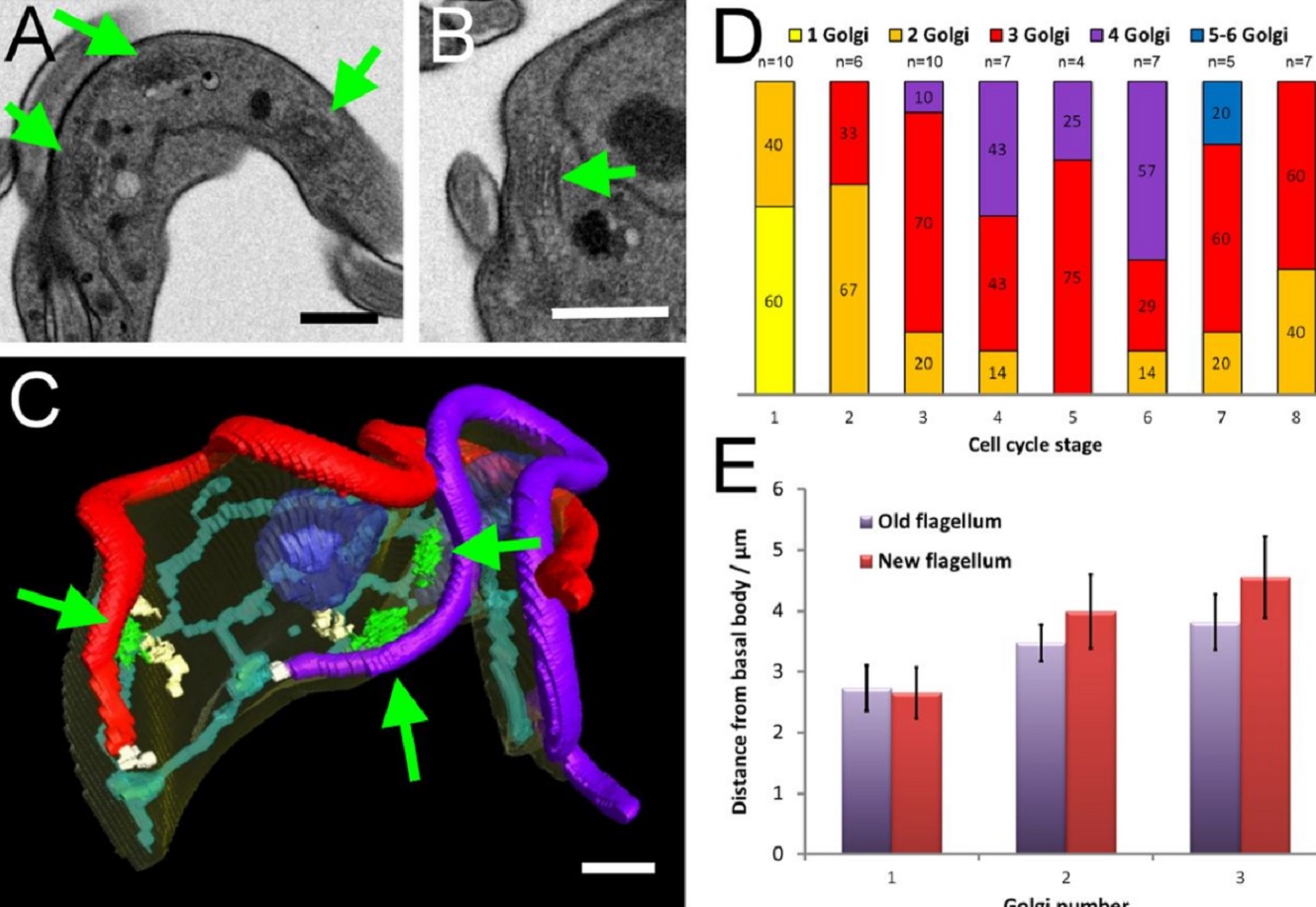
The major mammalian bloodstream form of the African sleeping sickness parasite Trypanosoma bruceimultiplies rapidly, and it is important to understand how these cells divide. Organelle inheritance involves complex spatiotemporal re-arrangements to ensure correct distribution to daughter cells…
How Amira-Avizo Software is used
Segmentation was conducted using AMIRA (versions 5.3.3 to 6.1, FEI, Eindhoven). Whole cells were segmented by selecting a region of interest and applying a threshold based on the density of stain. Manual segmentation of organelles was conducted using the brush tool and was based on ultrastructural characteristics specific to each organelle. Each organelle was segmented using the organelle membrane as the outer edge. Surface models were produced for each segmented organelle. The movies shown in the supplementary material were generated using the demo-maker and movie-maker functions in AMIRA and the 3D measurement tool was used to take measurements. Distance measurements were taken from similar points in 3D space between organelles to enable an accurate comparison across the cell cycle, for example, the most posterior point of the nucleus was always used. When measuring distances between organelles, the most posterior point of each organelle was used as the start and end point of measurement, for example, between the basal bodies and Golgi bodies. Volumes of organelles were automatically generated from the surface renderings of segmented data. The volume of the nuclei included the nucleolus. The volume of the kinetoplast only included the stained DNA region, not the surrounding mitochondrial matrix or membrane.